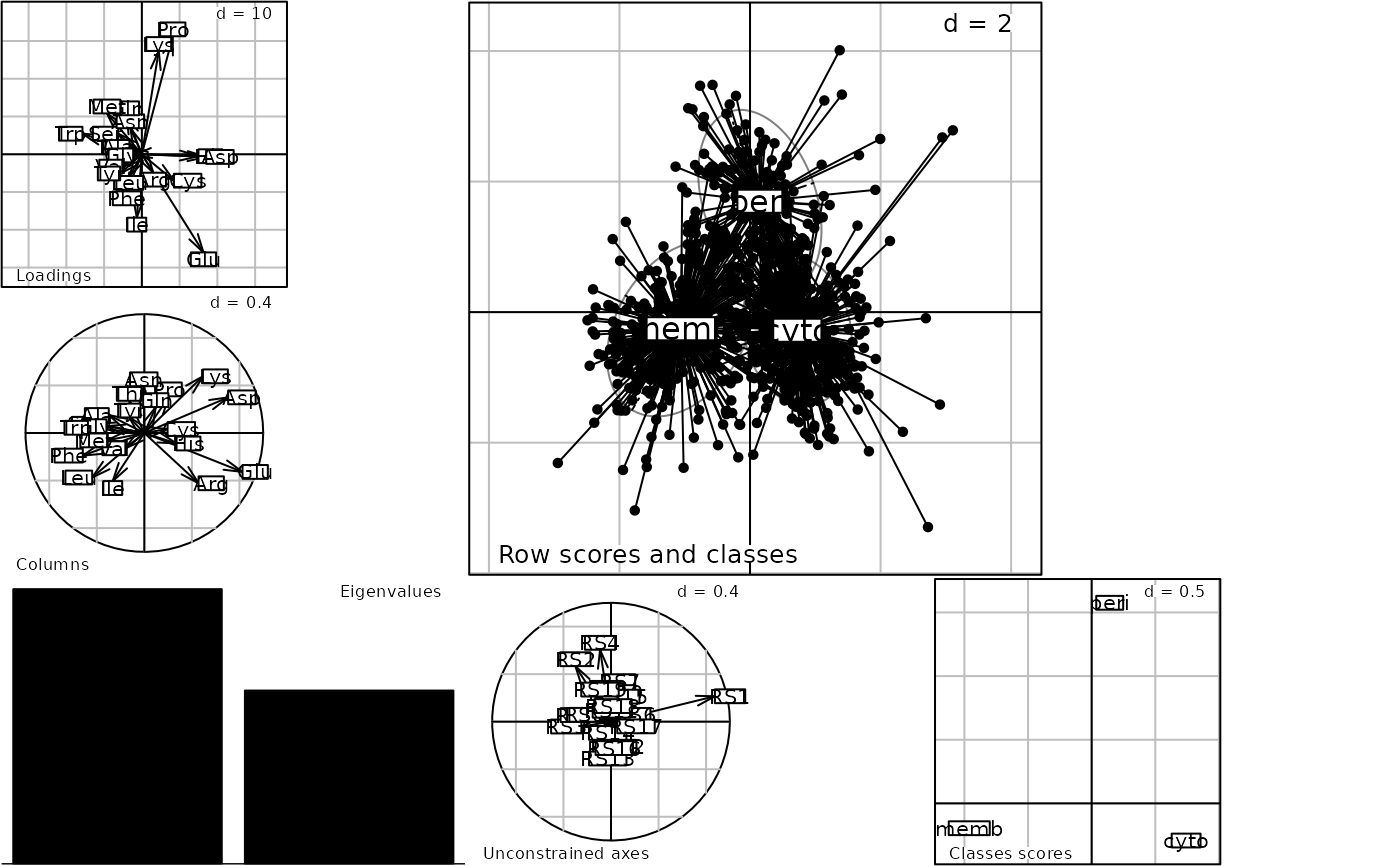
Discriminant Correspondence Analysis
discrimin.coa.Rdperforms a discriminant correspondence analysis.
Value
a list of class discrimin. See discrimin
References
Perriere, G.,Lobry, J. R. and Thioulouse J. (1996) Correspondence discriminant analysis: a multivariate method for comparing
classes of protein and nucleic acid sequences. CABIOS, 12, 519–524.
Perriere, G. and Thioulouse, J. (2003) Use of Correspondence Discriminant Analysis to predict the subcellular location of bacterial proteins. Computer Methods and Programs in Biomedicine, 70, 2, 99–105.
Author
Daniel Chessel
Anne-Béatrice Dufour anne-beatrice.dufour@univ-lyon1.fr