Phylogeny and quantitative traits of flowers
tithonia.RdThis data set describes the phylogeny of 11 flowers as reported by Morales (2000). It also gives morphologic and demographic traits corresponding to these 11 species.
Usage
data(tithonia)Format
tithonia is a list containing the 2 following objects :
- tre
is a character string giving the phylogenetic tree in Newick format.
- tab
is a data frame with 11 species and 14 traits (6 morphologic traits and 8 demographic).
Details
Variables of tithonia$tab are the following ones :
morho1: is a numeric vector that describes the seed size (mm)
morho2: is a numeric vector that describes the flower size (mm)
morho3: is a numeric vector that describes the female leaf size (cm)
morho4: is a numeric vector that describes the head size (mm)
morho5: is a integer vector that describes the number of flowers per head
morho6: is a integer vector that describes the number of seeds per head
demo7: is a numeric vector that describes the seedling height (cm)
demo8: is a numeric vector that describes the growth rate (cm/day)
demo9: is a numeric vector that describes the germination time
demo10: is a numeric vector that describes the establishment (per cent)
demo11: is a numeric vector that describes the viability (per cent)
demo12: is a numeric vector that describes the germination (per cent)
demo13: is a integer vector that describes the resource allocation
demo14: is a numeric vector that describes the adult height (m)
Source
Data were obtained from Morales, E. (2000) Estimating phylogenetic inertia in Tithonia (Asteraceae) : a comparative approach. Evolution, 54, 2, 475–484.
Examples
data(tithonia)
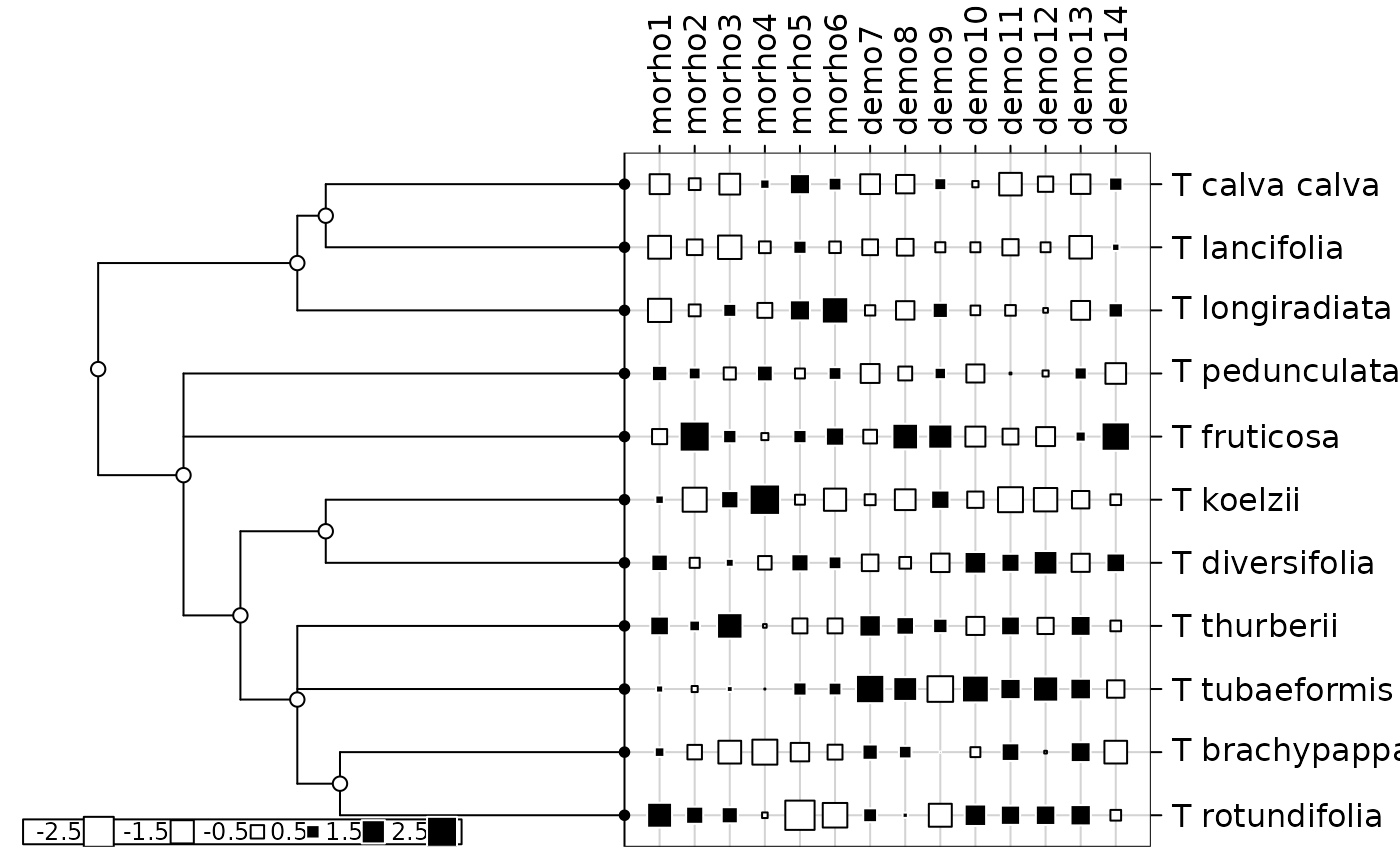
phy <- newick2phylog(tithonia$tre)
tab <- log(tithonia$tab + 1)
table.phylog(scalewt(tab), phy)
 gearymoran(phy$Wmat, tab)
#> class: krandtest lightkrandtest
#> Monte-Carlo tests
#> Call: as.krandtest(sim = matrix(res$result, ncol = nvar, byrow = TRUE),
#> obs = res$obs, alter = alter, names = test.names)
#>
#> Number of tests: 14
#>
#> Adjustment method for multiple comparisons: none
#> Permutation number: 999
#> Test Obs Std.Obs Alter Pvalue
#> 1 morho1 0.7321356 4.4756496 greater 0.004
#> 2 morho2 0.3822949 0.3431711 greater 0.323
#> 3 morho3 0.3712126 0.3186884 greater 0.261
#> 4 morho4 0.2572795 -0.9215946 greater 0.783
#> 5 morho5 0.4457180 1.1775848 greater 0.118
#> 6 morho6 0.4089212 0.8161762 greater 0.161
#> 7 demo7 0.4416215 1.1715011 greater 0.092
#> 8 demo8 0.4822195 1.7040732 greater 0.066
#> 9 demo9 0.3043863 -0.4368997 greater 0.623
#> 10 demo10 0.2744296 -0.7745213 greater 0.809
#> 11 demo11 0.4458932 1.2905809 greater 0.105
#> 12 demo12 0.2640213 -0.9029662 greater 0.863
#> 13 demo13 0.6092138 3.5006374 greater 0.004
#> 14 demo14 0.3903768 0.4925647 greater 0.243
#>
gearymoran(phy$Amat, tab)
#> class: krandtest lightkrandtest
#> Monte-Carlo tests
#> Call: as.krandtest(sim = matrix(res$result, ncol = nvar, byrow = TRUE),
#> obs = res$obs, alter = alter, names = test.names)
#>
#> Number of tests: 14
#>
#> Adjustment method for multiple comparisons: none
#> Permutation number: 999
#> Test Obs Std.Obs Alter Pvalue
#> 1 morho1 0.53784586 2.92757199 greater 0.006
#> 2 morho2 0.10046720 0.59542278 greater 0.276
#> 3 morho3 0.07014773 0.40140646 greater 0.325
#> 4 morho4 -0.09223746 -0.59479431 greater 0.767
#> 5 morho5 0.35119692 2.18143858 greater 0.026
#> 6 morho6 0.17659490 1.12223812 greater 0.143
#> 7 demo7 0.44981278 2.61801829 greater 0.010
#> 8 demo8 0.25528857 1.44887928 greater 0.085
#> 9 demo9 -0.01050264 -0.08490146 greater 0.494
#> 10 demo10 -0.09660405 -0.57036811 greater 0.674
#> 11 demo11 0.26985310 1.42718089 greater 0.094
#> 12 demo12 -0.19861554 -1.13662673 greater 0.875
#> 13 demo13 0.63572312 3.35296073 greater 0.003
#> 14 demo14 0.05085783 0.31633099 greater 0.360
#>
gearymoran(phy$Wmat, tab)
#> class: krandtest lightkrandtest
#> Monte-Carlo tests
#> Call: as.krandtest(sim = matrix(res$result, ncol = nvar, byrow = TRUE),
#> obs = res$obs, alter = alter, names = test.names)
#>
#> Number of tests: 14
#>
#> Adjustment method for multiple comparisons: none
#> Permutation number: 999
#> Test Obs Std.Obs Alter Pvalue
#> 1 morho1 0.7321356 4.4756496 greater 0.004
#> 2 morho2 0.3822949 0.3431711 greater 0.323
#> 3 morho3 0.3712126 0.3186884 greater 0.261
#> 4 morho4 0.2572795 -0.9215946 greater 0.783
#> 5 morho5 0.4457180 1.1775848 greater 0.118
#> 6 morho6 0.4089212 0.8161762 greater 0.161
#> 7 demo7 0.4416215 1.1715011 greater 0.092
#> 8 demo8 0.4822195 1.7040732 greater 0.066
#> 9 demo9 0.3043863 -0.4368997 greater 0.623
#> 10 demo10 0.2744296 -0.7745213 greater 0.809
#> 11 demo11 0.4458932 1.2905809 greater 0.105
#> 12 demo12 0.2640213 -0.9029662 greater 0.863
#> 13 demo13 0.6092138 3.5006374 greater 0.004
#> 14 demo14 0.3903768 0.4925647 greater 0.243
#>
gearymoran(phy$Amat, tab)
#> class: krandtest lightkrandtest
#> Monte-Carlo tests
#> Call: as.krandtest(sim = matrix(res$result, ncol = nvar, byrow = TRUE),
#> obs = res$obs, alter = alter, names = test.names)
#>
#> Number of tests: 14
#>
#> Adjustment method for multiple comparisons: none
#> Permutation number: 999
#> Test Obs Std.Obs Alter Pvalue
#> 1 morho1 0.53784586 2.92757199 greater 0.006
#> 2 morho2 0.10046720 0.59542278 greater 0.276
#> 3 morho3 0.07014773 0.40140646 greater 0.325
#> 4 morho4 -0.09223746 -0.59479431 greater 0.767
#> 5 morho5 0.35119692 2.18143858 greater 0.026
#> 6 morho6 0.17659490 1.12223812 greater 0.143
#> 7 demo7 0.44981278 2.61801829 greater 0.010
#> 8 demo8 0.25528857 1.44887928 greater 0.085
#> 9 demo9 -0.01050264 -0.08490146 greater 0.494
#> 10 demo10 -0.09660405 -0.57036811 greater 0.674
#> 11 demo11 0.26985310 1.42718089 greater 0.094
#> 12 demo12 -0.19861554 -1.13662673 greater 0.875
#> 13 demo13 0.63572312 3.35296073 greater 0.003
#> 14 demo14 0.05085783 0.31633099 greater 0.360
#>