Originality of a species
originality.Rdcomputes originality values for species from an ultrametric phylogenetic tree.
Details
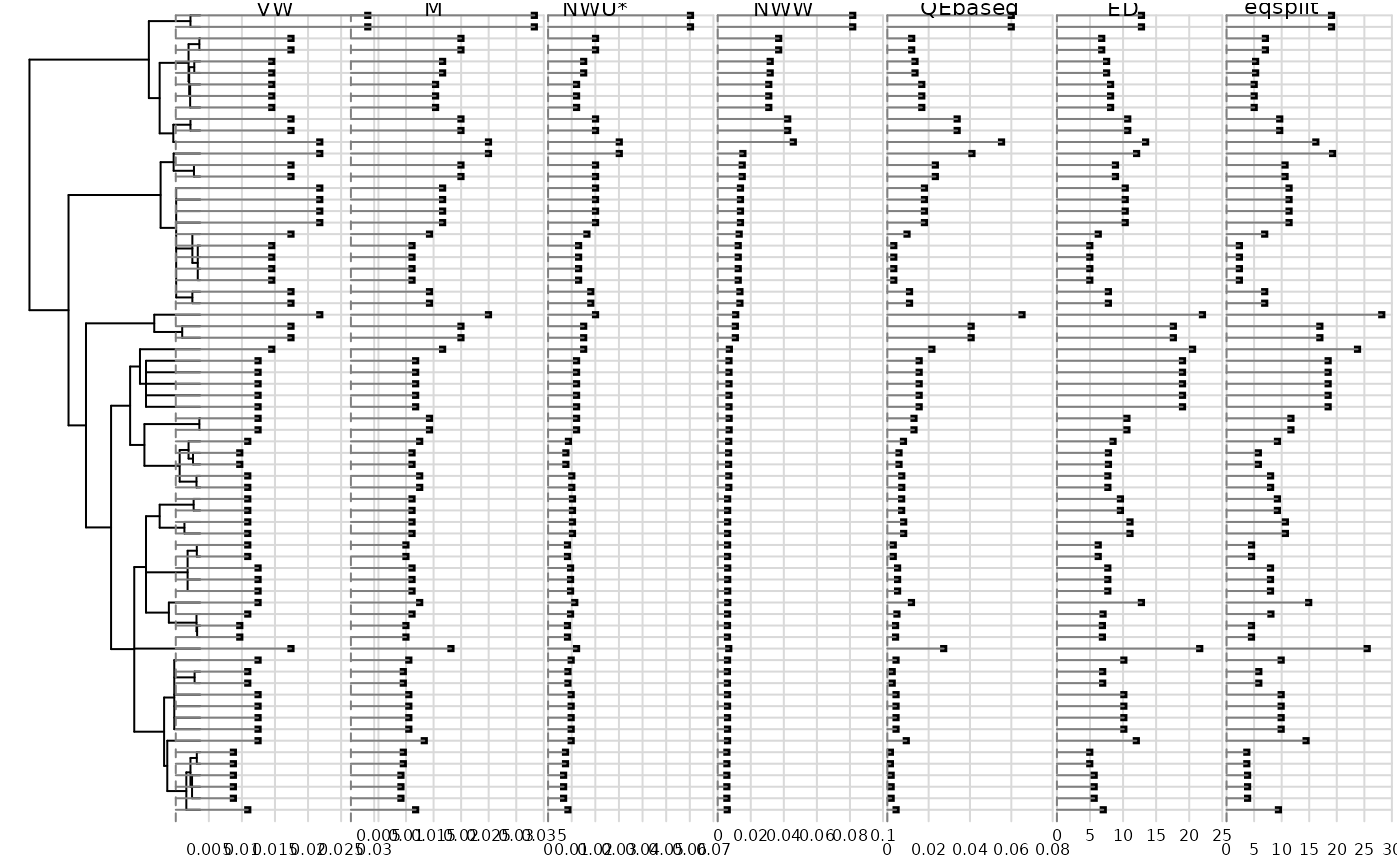
1 = Vane-Wright et al.'s (1991) node-counting index 2 = May's (1990) branch-counting index 3 = Nixon and Wheeler's (1991) unweighted index, based on the sum of units in binary values 4 = Nixon and Wheeler's (1991) weighted index 5 = QE-based index 6 = Isaac et al. (2007) ED index 7 = Redding et al. (2006) Equal-split index
Value
Returns a data frame with species in rows, and the selected indices of originality in columns. Indices are expressed as percentages.
References
Isaac, N.J.B., Turvey, S.T., Collen, B., Waterman, C. and Baillie, J.E.M. (2007) Mammals on the EDGE: conservation priorities based on threat and phylogeny. PloS ONE, 2, e–296.
Redding, D. and Mooers, A. (2006) Incorporating evolutionary measures into conservation prioritization. Conservation Biology, 20, 1670–1678.
Pavoine, S., Ollier, S. and Dufour, A.-B. (2005) Is the originality of a species measurable? Ecology Letters, 8, 579–586.
Vane-Wright, R.I., Humphries, C.J. and Williams, P.H. (1991). What to protect? Systematics and the agony of choice. Biological Conservation, 55, 235–254.
May, R.M. (1990). Taxonomy as destiny. Nature, 347, 129–130.
Nixon, K.C. and Wheeler, Q.D. (1992). Measures of phylogenetic diversity. In: Extinction and Phylogeny (eds. Novacek, M.J. and Wheeler, Q.D.), 216–234, Columbia University Press, New York.
Author
Sandrine Pavoine pavoine@mnhn.fr
Examples
data(carni70)
carni70.phy <- newick2phylog(carni70$tre)
ori.tab <- originality(carni70.phy, 1:7)
names(ori.tab)
#> [1] "VW" "M" "NWU*" "NWW" "QEbased" "ED" "eqsplit"
dotchart.phylog(carni70.phy, ori.tab, scaling = FALSE, yjoining = 0,
ranging = FALSE, cleaves = 0, ceti = 0.5, csub = 0.7, cdot = 0.5)