Mantel test (correlation between two distance matrices (in R).)
mantel.rtest.RdPerforms a Mantel test between two distance matrices.
References
Mantel, N. (1967) The detection of disease clustering and a generalized regression approach. Cancer Research, 27, 209–220.
Author
Daniel Chessel
Stéphane Dray stephane.dray@univ-lyon1.fr
Examples
data(yanomama)
gen <- quasieuclid(as.dist(yanomama$gen))
geo <- quasieuclid(as.dist(yanomama$geo))
plot(r1 <- mantel.rtest(geo,gen), main = "Mantel's test")
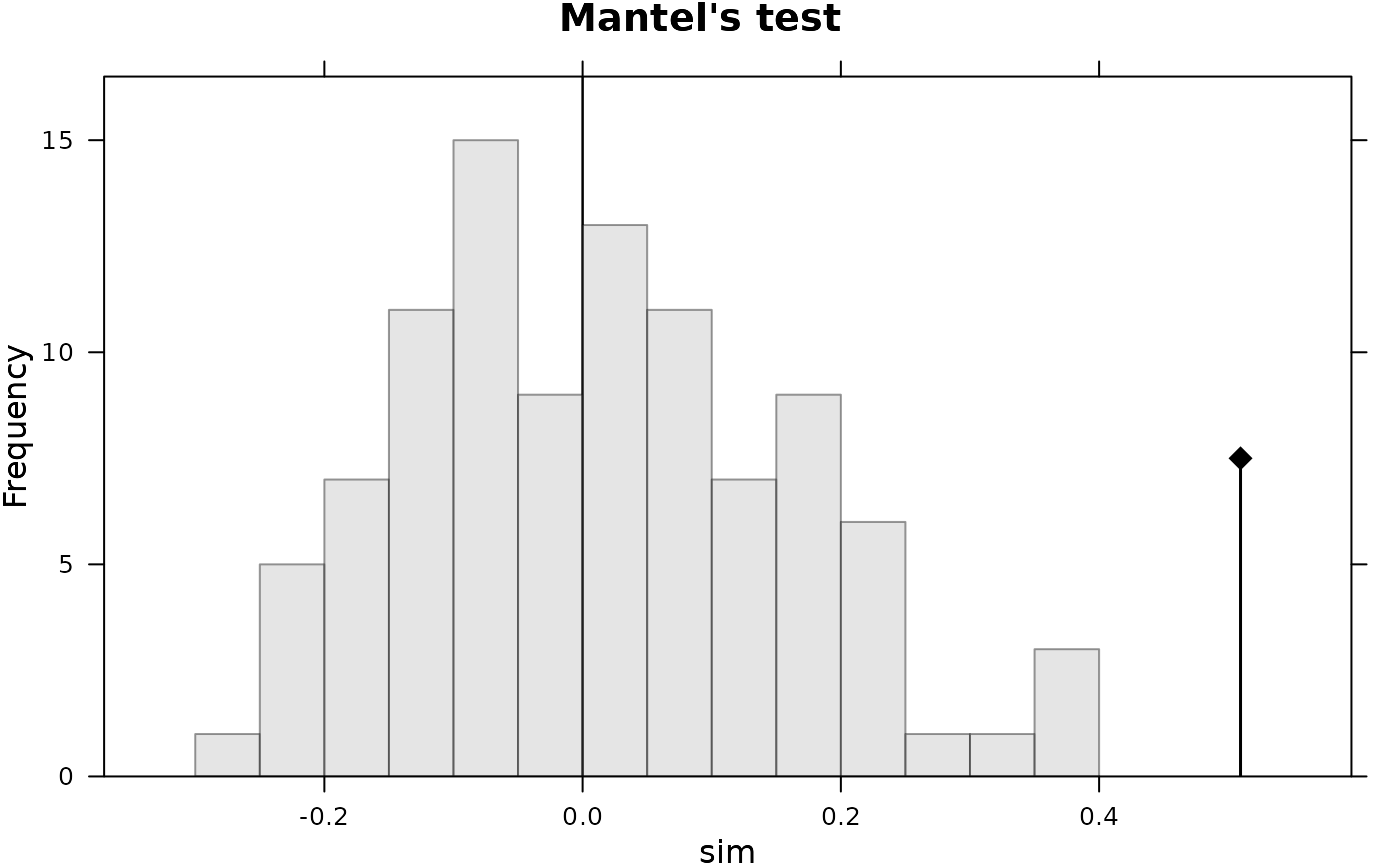 r1
#> Monte-Carlo test
#> Call: mantel.rtest(m1 = geo, m2 = gen)
#>
#> Observation: 0.5095199
#>
#> Based on 99 replicates
#> Simulated p-value: 0.01
#> Alternative hypothesis: greater
#>
#> Std.Obs Expectation Variance
#> 3.474064150 -0.005003676 0.021934863
r1
#> Monte-Carlo test
#> Call: mantel.rtest(m1 = geo, m2 = gen)
#>
#> Observation: 0.5095199
#>
#> Based on 99 replicates
#> Simulated p-value: 0.01
#> Alternative hypothesis: greater
#>
#> Std.Obs Expectation Variance
#> 3.474064150 -0.005003676 0.021934863