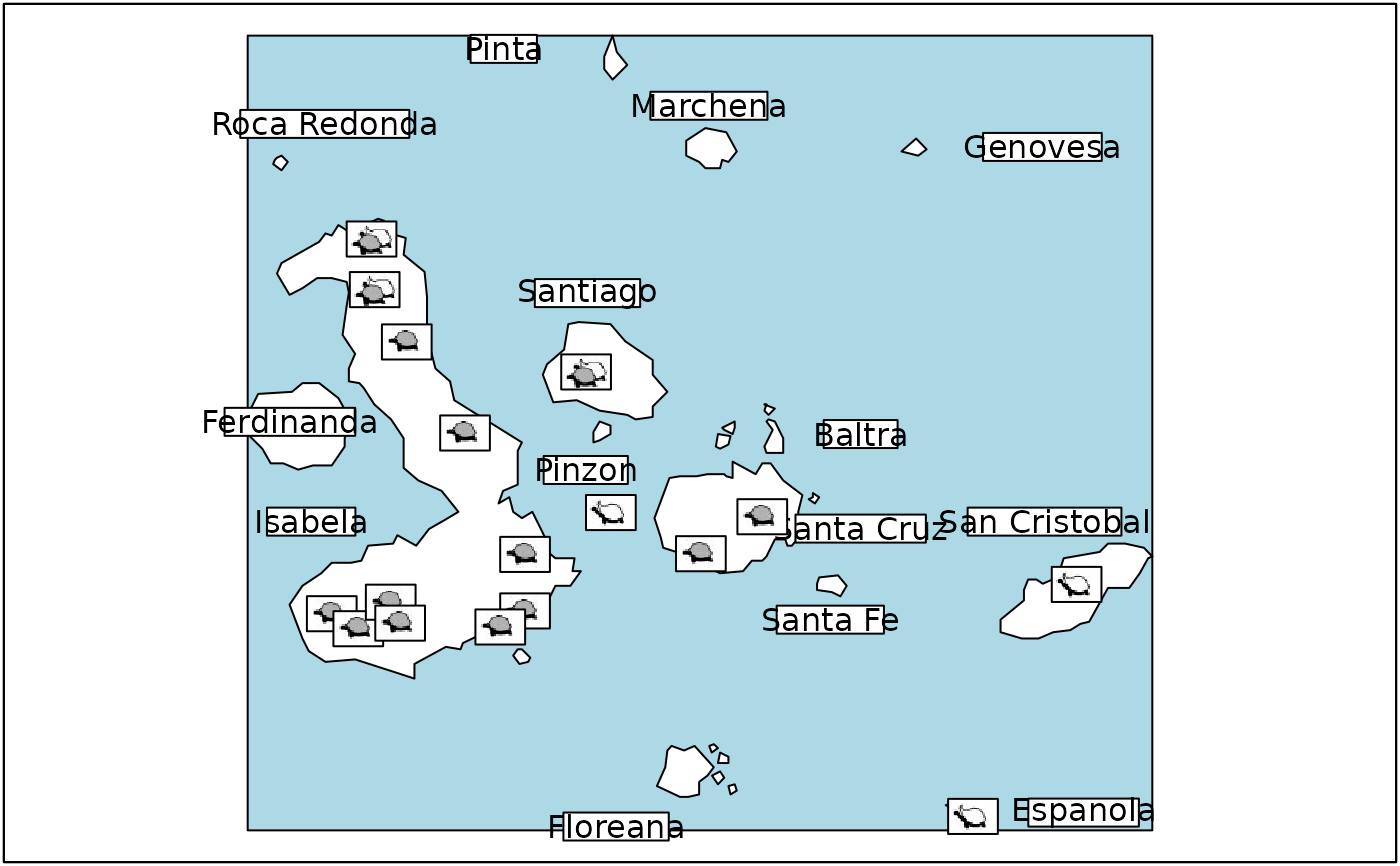
Microsatellites of Galapagos tortoises populations
ggtortoises.RdThis data set gives genetic relationships between Galapagos tortoises populations with 10 microsatellites.
Usage
data(ggtortoises)Format
ggtortoises is a list with the following components:
- area
a data frame designed to be used in the
area.plotfunction- ico
a list of three pixmap icons representing the tortoises morphotypes
- pop
a data frame containing meta informations about populations
- misc
a data frame containing the coordinates of the island labels
- loc
a numeric vector giving the number of alleles by marker
- tab
a data frame containing the number of alleles by populations for 10 microsatellites
- Spatial
an object of the class
SpatialPolygonsofsp, containing the map
Source
M.C. Ciofi, C. Milinkovitch, J.P. Gibbs, A. Caccone, and J.R. Powell (2002) Microsatellite analysis of genetic divergence among populations of giant galapagos tortoises. Molecular Ecology 11: 2265-2283.
References
M.C. Ciofi, C. Milinkovitch, J.P. Gibbs, A. Caccone, and J.R. Powell (2002). Microsatellite analysis of genetic divergence among populations of giant galapagos tortoises. Molecular Ecology 11: 2265-2283.
See a data description at http://pbil.univ-lyon1.fr/R/pdf/pps069.pdf (in French).
Examples
if(requireNamespace("pixmap", quietly=TRUE)) {
data(ggtortoises)
if(adegraphicsLoaded()) {
if(requireNamespace("sp", quietly = TRUE)) {
g1 <- s.logo(ggtortoises$pop, ggtortoises$ico[as.character(ggtortoises$pop$carap)],
Sp = ggtortoises$Spatial, pbackground.col = "lightblue", pSp.col = "white",
pgrid.draw = FALSE, ppoints.cex = 0.5)
g1 <- s.label(ggtortoises$misc, pgrid.draw = FALSE, porigin.include = FALSE,
paxes.draw = FALSE, add = TRUE)
}
} else {
a1 <- ggtortoises$area
area.plot(a1)
rect(min(a1$x), min(a1$y), max(a1$x), max(a1$y), col = "lightblue")
invisible(lapply(split(a1, a1$id), function(x) polygon(x[, -1], col = "white")))
s.label(ggtortoises$misc, grid = FALSE, include.ori = FALSE, addaxes = FALSE, add.p = TRUE)
listico <- ggtortoises$ico[as.character(ggtortoises$pop$carap)]
s.logo(ggtortoises$pop, listico, add.p = TRUE)
}
}