Phylogenies and quantitative traits from Abouheif
abouheif.eg.RdThis data set gathers three phylogenies with three sets of traits as reported by Abouheif (1999).
Usage
data(abouheif.eg)Format
abouheif.eg is a list containing the 6 following objects :
- tre1
is a character string giving the first phylogenetic tree made up of 8 leaves.
- vec1
is a numeric vector with 8 values.
- tre2
is a character string giving the second phylogenetic tree made up of 7 leaves.
- vec2
is a numeric vector with 7 values.
- tre3
is a character string giving the third phylogenetic tree made up of 15 leaves.
- vec3
is a numeric vector with 15 values.
References
Abouheif, E. (1999) A method for testing the assumption of phylogenetic independence in comparative data. Evolutionary Ecology Research, 1, 895–909.
Examples
data(abouheif.eg)
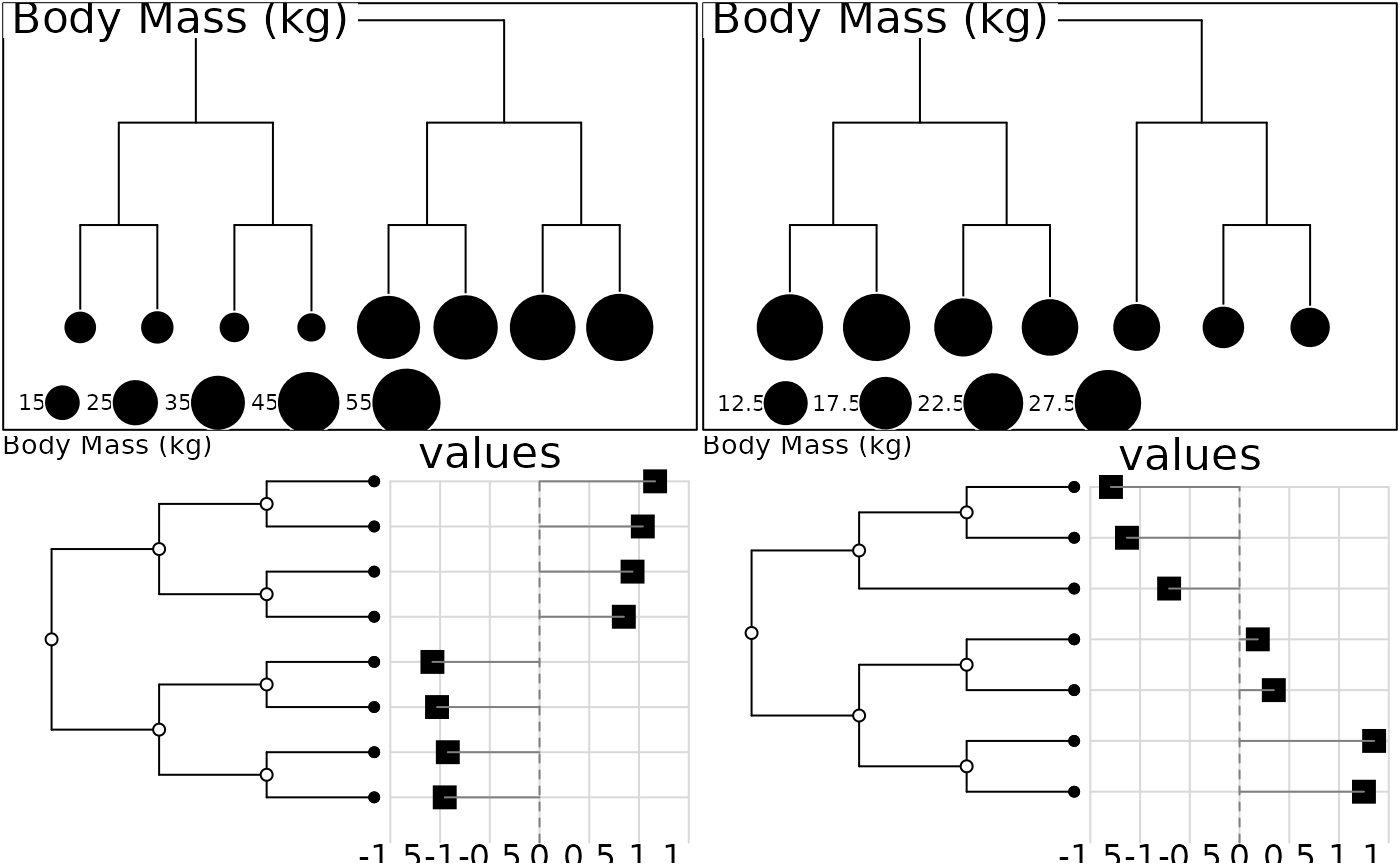
par(mfrow=c(2,2))
symbols.phylog(newick2phylog(abouheif.eg$tre1), abouheif.eg$vec1,
sub = "Body Mass (kg)", csi = 2, csub = 2)
symbols.phylog(newick2phylog(abouheif.eg$tre2), abouheif.eg$vec2,
sub = "Body Mass (kg)", csi = 2, csub = 2)
dotchart.phylog(newick2phylog(abouheif.eg$tre1), abouheif.eg$vec1,
sub = "Body Mass (kg)", cdot = 2, cnod = 1, possub = "topleft",
csub = 2, ceti = 1.5)
dotchart.phylog(newick2phylog(abouheif.eg$tre2), abouheif.eg$vec2,
sub = "Body Mass (kg)", cdot = 2, cnod = 1, possub = "topleft",
csub = 2, ceti = 1.5)
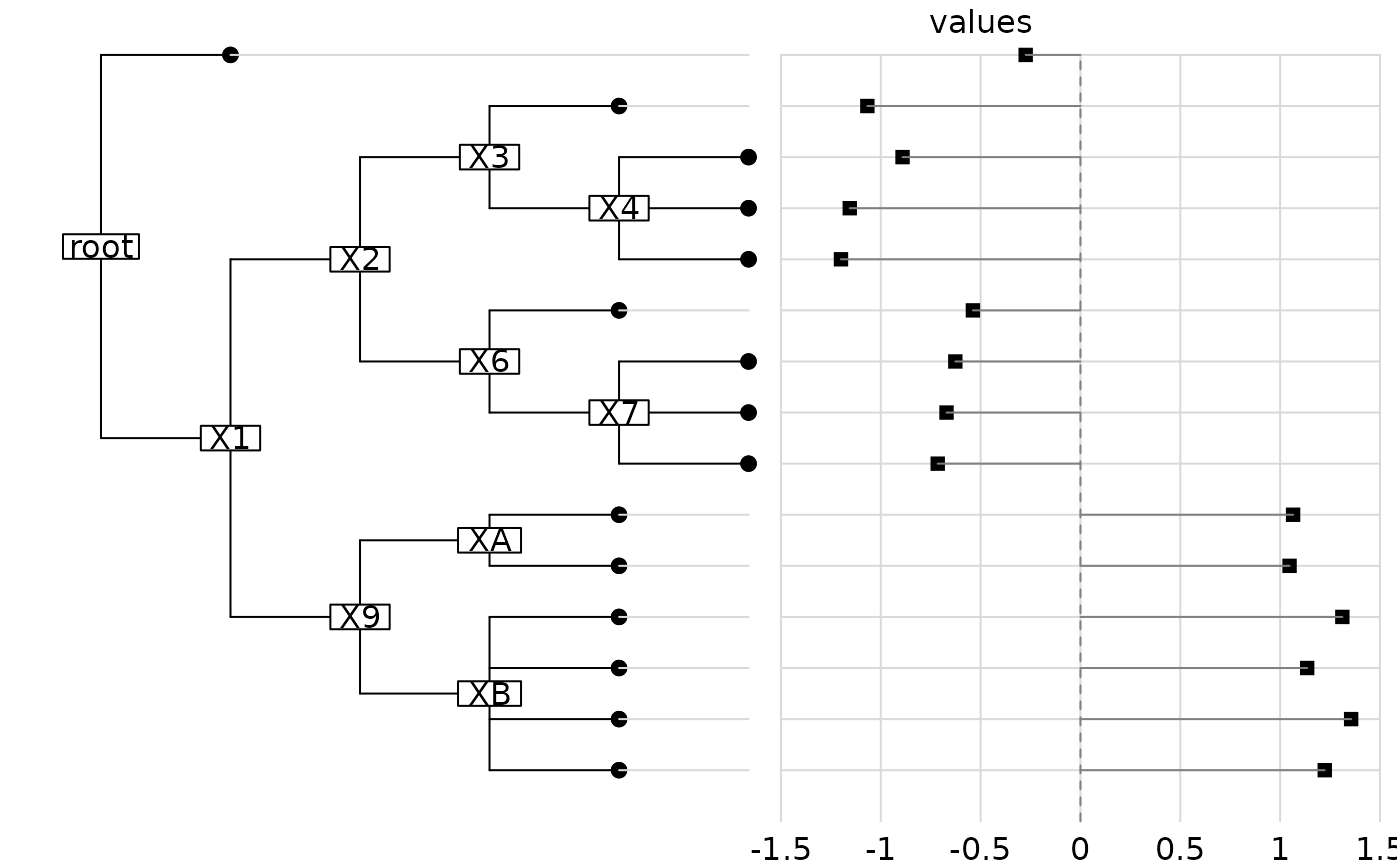
 par(mfrow = c(1,1))
w.phy=newick2phylog(abouheif.eg$tre3)
dotchart.phylog(w.phy,abouheif.eg$vec3, clabel.n = 1)
par(mfrow = c(1,1))
w.phy=newick2phylog(abouheif.eg$tre3)
dotchart.phylog(w.phy,abouheif.eg$vec3, clabel.n = 1)