Compute a multivariate empirical variogram. It is strictly equivalent to summing univariate variograms
Arguments
- Y
A matrix with numeric data
- xy
A matrix with coordinates of samples
- dmin
The minimum distance value at which the variogram is computed (i.e. lower bound of the first class)
- dmax
The maximum distance value at which the variogram is computed (i.e. higher bound of the last class)
- nclass
Number of classes of distances
Value
A list:
- d
Distances (i.e. centers of distance classes).
- var
Empirical semi-variances.
- n.w
Number of connections between samples for a given distance.
- n.c
Number of samples used for the computation of the variogram.
- dclass
Character vector with the names of the distance classes.
References
Wagner H. H. (2003) Spatial covariance in plant communities: integrating ordination, geostatistics, and variance testing. Ecology, 84, 1045–1057
Examples
if(require(ade4)){
data(oribatid)
# Hellinger transformation
fau <- sqrt(oribatid$fau / outer(apply(oribatid$fau, 1, sum), rep(1, ncol(oribatid$fau)), "*"))
# Removing linear effect
faudt <- resid(lm(as.matrix(fau) ~ as.matrix(oribatid$xy)))
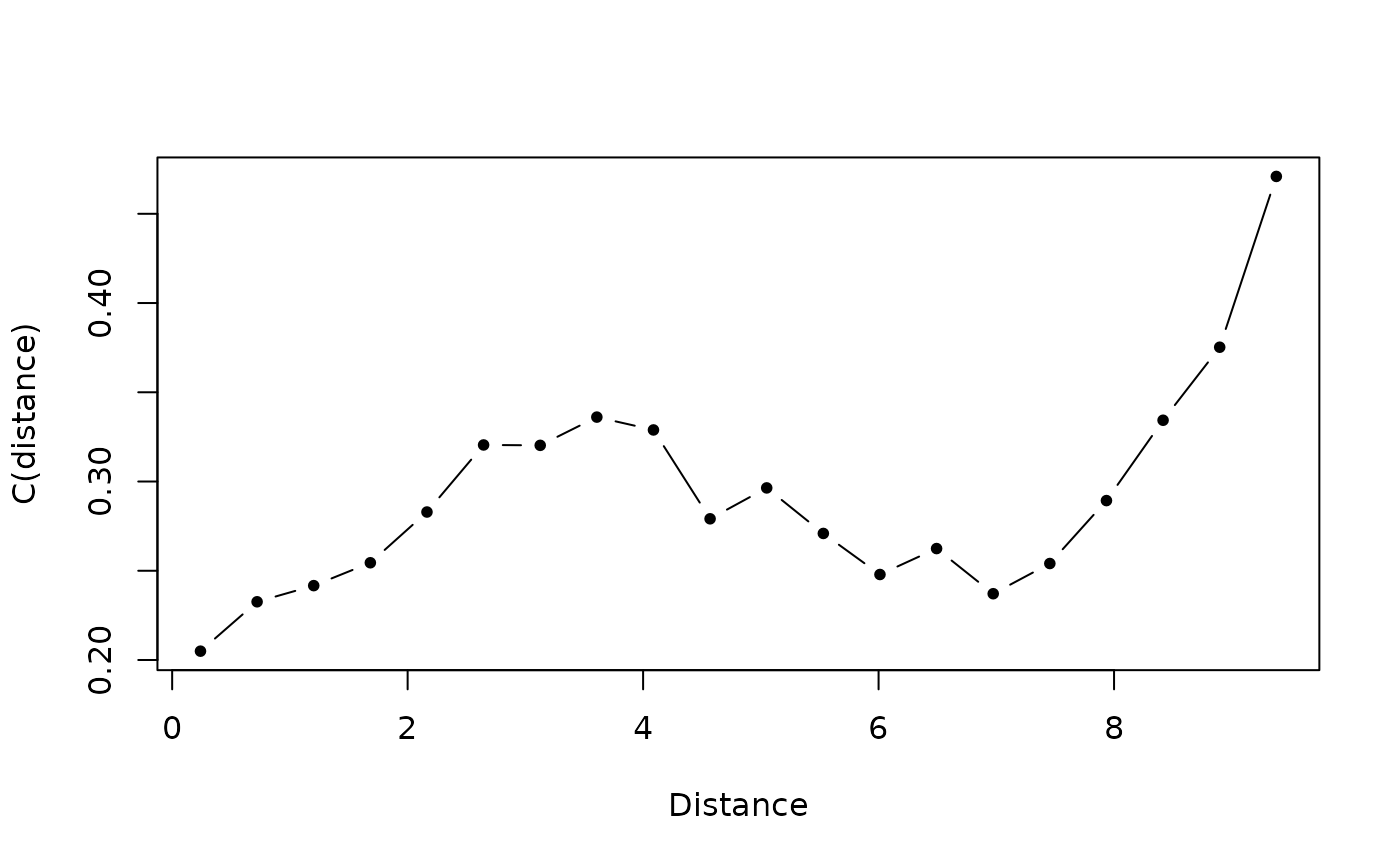
mvspec <- variogmultiv(faudt, oribatid$xy, nclass = 20)
mvspec
plot(mvspec$d, mvspec$var,type = 'b', pch = 20, xlab = "Distance", ylab = "C(distance)")
}