Perform a test of the shared space-environment fraction of a variation partitioning using torus-translation (TT) or Moran Spectral Randomisation (MSR)
Source:R/envspace.test.R
envspace.test.RdThe function uses two different spatially-constrained null models to test the shared space-environment fraction (SSEF, or fraction [b]) of a variation partitioning of two explanatory components.
envspace.test(
spe,
env,
coord,
MEM.spe,
listw.env,
MEM.autocor = c("positive", "negative", "all"),
regular = FALSE,
nperm = 999,
MSR.method = "singleton",
alpha = 0.05
)Arguments
- spe
Vector, matrix, or dataframe of response variable(s) (e.g. species abundances)
- env
Vector, matrix, or dataframe of environmental variables (rows = sites, columns = variables)
- coord
Matrix or dataframe of spatial coordinates of the sampled sites
- MEM.spe
Matrix or dataframe of spatial predictors (MEM variables) selected for
spe- listw.env
An object of class
listw(spatial weights) created by the functions of thespdeppackage or returned bylistw.candidates- MEM.autocor
A string indicating the type of spatial structure of interest for
env("positive","negative", or"all", for positive, negative, or both types of spatial autocorrelations, respectively); Default is"positive"- regular
Logical argument indicating whether a torus-translation test will be performed, in addition to the MSR. Set to
TRUEonly if the sampling design is regular (same number of sites on each line, same number of sites on each column). Set toFALSEotherwise; Default isFALSE- nperm
Number of permutations performed; Default is 999
- MSR.method
Algorithm of
msrto be used to perform the MSR. The three available procedures are"singleton"(default),"pair", and"triplet"(seemsrfor details)- alpha
Threshold value of null hypothesis rejection for the test of a spatial structure in the environment, and for the shared environment-space fraction of the variation partitioning; Default is 0.05
Value
If the condition of env being spatially structured is fulfilled, the test
is performed and the function returns an object of class randtest containing the results of the test.
Details
The function tests the SSEF (also known as fraction [b]) of a variation
partitioning of a response variable or matrix (y) between an environmental and a
spatial component (env, and MEM.spe, respectively). The SSEF is the
explained variation of y shared by env and MEM.spe.
The adjusted R-squared (Peres-Neto et al. 2006; R2adj) of the SSEF is not an
actual R2, as it is computed by subtracting the adjusted R2adj of other fractions and
therefore has zero degree of freedom (Legendre and Legendre 2012).
The SSEF can therefore not be computed in the classical way (residuals permutation;
Anderson and Legendre 1999, Legendre and Legendre 2012).
The function envspace.test provides two ways of testing this fraction, that is,
spatially-constrained null models based either on a torus-translation test (TT) (for
regular sampling designs only), or on Moran spectral randomizations (MSR) (for any type
of sampling design). The test of the SSEF should only be performed if both the global
models of y against all the environmental variables and against all spatial variables
are significant (see Bauman et al. 2018c).
The function first checks whether the environment displays significant spatial structures,
and then proceeds to the test of the SSEF if this condition is fulfilled (details in
Bauman et al. 2018c).
spe can be a vector or a multicolumn matrix or dataframe (multivariate
response data). If multivariate, it is greatly advised to transform spe prior
to performing the variation partitioning and testing the SSEF (e.g., Hellinger
transformation; see Legendre and Gallagher 2001).
MEM.spe is a set of spatial predictors (MEM variables). It is recommended to be
a well-defined subset of MEM variables selected among the complete set generated from
the spatial weighting matrix (SWM) (see review about spatial eigenvector selection in
Bauman et al. 2018a).
Optimising the selection of a subset of forward-selected MEM variables
among a set of candidate SWMs has been shown to increase statistical power as well as
R2-estimation accuracy (Bauman et al. 2018b). To do so, MEM.spe can be generated
using listw.candidates followed by listw.select. If a SWM has
already been selected in another way, then mem.select can be used to
generate the MEM variables and to select an optimal subset among them, which can then
be used as MEM.spe in envspace.test (see Details of function
mem.select).
listw.env corresponds to the SWM that will be used to test for a spatial structure
in env, and to build the MEM variables for the MSR test.
The choice of the SWM for env can also be optimised with listw.select.
The SWMs selected for spe and env should be optimised separately to
best model the spatial structure of both spe and env (see example).
To verify that env displays a significant spatial pattern, prior to performing the
test of the SSEF, a residuals permutation test is performed on the global set of MEM
variables (generated internally from listw.env) associated to the type of
spatial structure of interest (see argument MEM.autocor). This test is performed
with mem.select. The choice of MEM.autocor should be made according to
the MEM.autocor argument used to build MEM.spe.
env is a dataset of environmental variables chosen by the user. We recommend dealing
with collinearity issues prior to performing the variation partitioning and the test of
the SSEF (see Dormann et al. 2013 for a review of methods to cope with collinearity).
regular is a logical argument indicating whether a TT test should
be performed instead of the MSR to test the SSEF. Since the TT can only
be performed on regular sampling designs, regular should only be set to
TRUE if the sampling design is either a transect, or a grid displaying the
same number of sites for all lines and columns (although the number of sites per column
can differ from the number of sites per line).
listw.env is the SWM used by the MSR to generate spatially-constrained null
environmental variables. It should ideally be a SWM optimised on the basis of env
using the function listw.select, with the argument method = "global" (see
Details of function mem.select for an explanation).
This will allow detecting the spatial structures of env as accurately as possible,
hence allowing MSR to generate null environmental variables as spatially faithful to the
original ones.
It is also on the basis of listw.env that MEM variables will be generated to test
whether env is spatially structured (i.e. global test) prior to perform the test of
the SSEF.
It is worth mentioning that, although a significant SSEF may provide evidence of an induced spatial dependence (Bauman et al. 2018c), a non-significant SSEF only indicates that no induced spatial dependence could be detected in relation with the chosen environmental variables. This does not exclude that this effect may exist with respect to some unmeasured variables.
References
Anderson M. and Legendre P. (1999) An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. Journal of Statistical Computation and Simulation, 62(3), 271–303
Bauman D., Drouet T., Dray S. and Vleminckx J. (2018a) Disentangling good from bad practices in the selection of spatial or phylogenetic eigenvectors. Ecography, 41, 1–12
Bauman D., Fortin M-J, Drouet T. and Dray S. (2018b) Optimizing the choice of a spatial weighting matrix in eigenvector-based methods. Ecology
Bauman D., Vleminckx J., Hardy O., Drouet T. (2018c) Testing and interpreting the shared space-environment fraction in variation partitioning analyses of ecological data. Oikos
Blanchet G., Legendre P. and Borcard D. (2008) Forward selection of explanatory variables. Ecology, 89(9), 2623–2632
Legendre P., Gallagher E.D. (2001) Ecologically meaningful transformations for ordination of species data. Oecologia, 129(2), 271–280
Legendre P. and Legendre L. (2012) Numerical Ecology, Elsevier, Amsterdam
Peres-Neto P., Legendre P., Dray S., Borcard D. (2006) Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology, 87(10), 2614–2625
Peres-Neto P. and Legendre P. (2010) Estimating and controlling for spatial structure in the study of ecological communities. Global Ecology and Biogeography, 19, 174–184
See also
Examples
# \donttest{
if(require(vegan)) {
# Illustration of the test of the SSEF on the oribatid mite data
# (Borcard et al. 1992, 1994 for details on the dataset):
# Community data (response matrix):
data(mite)
# Hellinger-transformation of the community data (Legendre and Gallagher 2001):
Y <- decostand(mite, method = "hellinger")
# Environmental explanatory dataset:
data(mite.env)
# We only use two numerical explanatory variables:
env <- mite.env[, 1:2]
dim(Y)
dim(env)
# Coordinates of the 70 sites:
data(mite.xy)
coord <- mite.xy
### Building a list of candidate spatial weighting matrices (SWMs) for the
### optimisation of the SWM selection, separately for 'Y' and 'env':
# We create five candidate SWMs: a connectivity matrix based on a Gabriel graphs, on
# a minimum spanning tree (i.e., two contrasted graph-based SWMs), either
# not weighted, or weighted by a linear function decreasing with the distance),
# and a distance-based SWM corresponding to the connectivity and weighting
# criteria of the original PCNM method:
candidates <- listw.candidates(coord, nb = c("gab", "mst", "pcnm"), weights = c("binary",
"flin"))
### Optimisation of the selection of a SWM:
# SWM for 'Y' (based on the best forward-selected subset of MEM variables):
modsel.Y <- listw.select(Y, candidates, method = "FWD", MEM.autocor = "positive",
p.adjust = TRUE)
names(candidates)[modsel.Y$best.id] # Best SWM selected
modsel.Y$candidates$Pvalue[modsel.Y$best.id] # Adjusted p-value of the global model
modsel.Y$candidates$N.var[modsel.Y$best.id] # Nb of forward-selected MEM variables
modsel.Y$candidates$R2Adj.select[modsel.Y$best.id] # Adjusted R2 of the selected MEM var.
# SWM for 'env' (method = "global" for the optimisation, as all MEM variables are required
# to use MSR):
modsel.env <- listw.select(env, candidates, method = "global", MEM.autocor = "positive",
p.adjust = TRUE)
names(candidates)[modsel.env$best.id] # Best SWM selected
modsel.env$candidates$Pvalue[modsel.env$best.id] # Adjusted p-value of the global model
modsel.env$candidates$N.var[modsel.env$best.id] # Nb of forward-selected MEM variables
modsel.env$candidates$R2Adj.select[modsel.env$best.id] # Adjusted R2 of the selected MEM var.
### We perform the variation partitioning:
# Subset of selected MEM variables within the best SWM:
MEM.spe <- modsel.Y$best$MEM.select
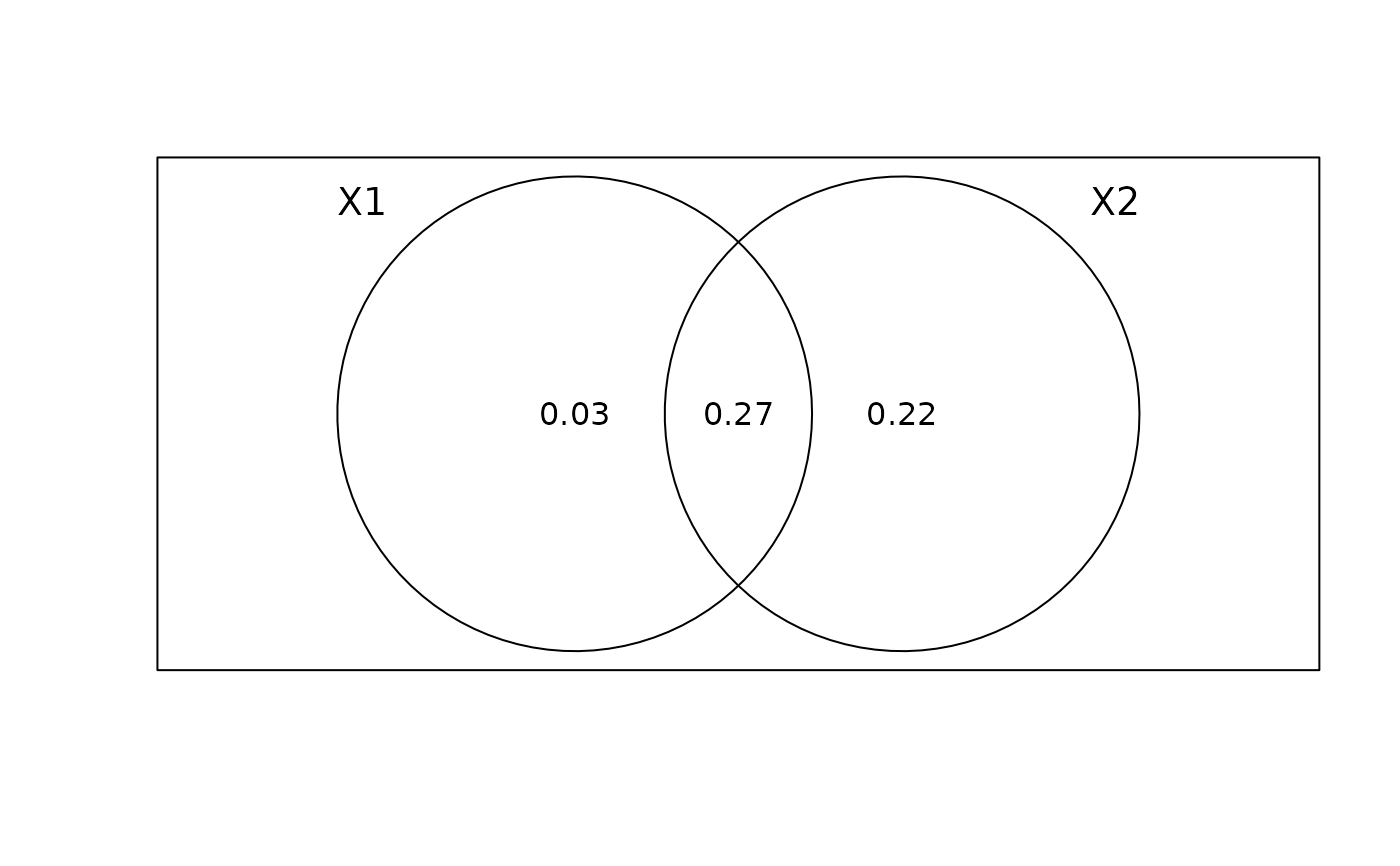
VP <- varpart(Y, env, MEM.spe)
plot(VP)
# Test of the shared space-environment fraction (fraction [b]):
SSEF.test <- envspace.test(Y, env, coord, MEM.spe,
listw.env = candidates[[modsel.env$best.id]],
regular = FALSE, nperm = 999)
SSEF.test
# The SSEF is highly significant, indicating a potential induced spatial dependence.
}
#> Warning: zero sum general weights
#> Procedure stopped (adjR2thresh criteria) adjR2cum = 0.452381 with 11 variables (> 0.446534)
#> Procedure stopped (alpha criteria): pvalue for variable 14 is 0.072000 (> 0.050000)
#> Procedure stopped (alpha criteria): pvalue for variable 14 is 0.098000 (> 0.050000)
#> Procedure stopped (alpha criteria): pvalue for variable 13 is 0.061000 (> 0.050000)
#> Procedure stopped (alpha criteria): pvalue for variable 12 is 0.061000 (> 0.050000)
 #> Monte-Carlo test
#> Call: as.randtest(sim = E.b, obs = R2.b, alter = alternative)
#>
#> Observation: 0.21545
#>
#> Based on 999 replicates
#> Simulated p-value: 1
#> Alternative hypothesis: greater
#>
#> Std.Obs Expectation Variance
#> -2.945501418 0.411513858 0.004430745
# }
#> Monte-Carlo test
#> Call: as.randtest(sim = E.b, obs = R2.b, alter = alternative)
#>
#> Observation: 0.21545
#>
#> Based on 999 replicates
#> Simulated p-value: 1
#> Alternative hypothesis: greater
#>
#> Std.Obs Expectation Variance
#> -2.945501418 0.411513858 0.004430745
# }