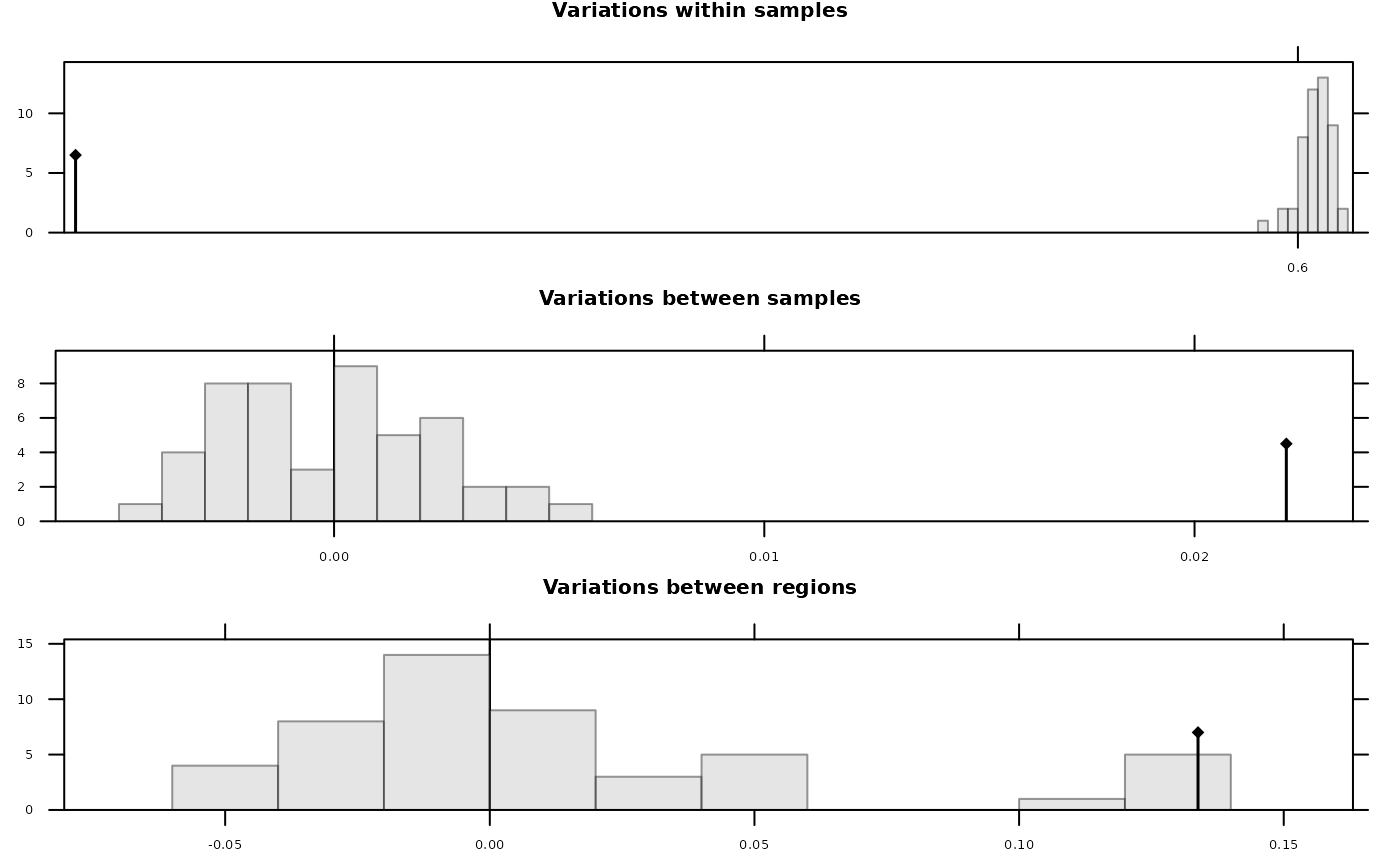
Permutation tests on an analysis of molecular variance (in C).
randtest.amova.RdTests the components of covariance with permutation processes described by Excoffier et al. (1992).
Usage
# S3 method for amova
randtest(xtest, nrepet = 99, ...)Arguments
- xtest
an object of class
amova- nrepet
the number of permutations
- ...
further arguments passed to or from other methods
References
Excoffier, L., Smouse, P.E. and Quattro, J.M. (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479--491.
Author
Sandrine Pavoine pavoine@mnhn.fr
Examples
data(humDNAm)
amovahum <- amova(humDNAm$samples, sqrt(humDNAm$distances), humDNAm$structures)
amovahum
#> $call
#> amova(samples = humDNAm$samples, distances = sqrt(humDNAm$distances),
#> structures = humDNAm$structures)
#>
#> $results
#> Df Sum Sq Mean Sq
#> Between regions 4 78.238115 19.5595288
#> Between samples Within regions 5 9.284744 1.8569488
#> Within samples 662 316.197379 0.4776395
#> Total 671 403.720238 0.6016695
#>
#> $componentsofcovariance
#> Sigma %
#> Variations Between regions 0.13380659 21.119144
#> Variations Between samples Within regions 0.02213345 3.493396
#> Variations Within samples 0.47763955 75.387459
#> Total variations 0.63357958 100.000000
#>
#> $statphi
#> Phi
#> Phi-samples-total 0.2461254
#> Phi-samples-regions 0.0442870
#> Phi-regions-total 0.2111914
#>
randtesthum <- randtest(amovahum, 49)
plot(randtesthum)